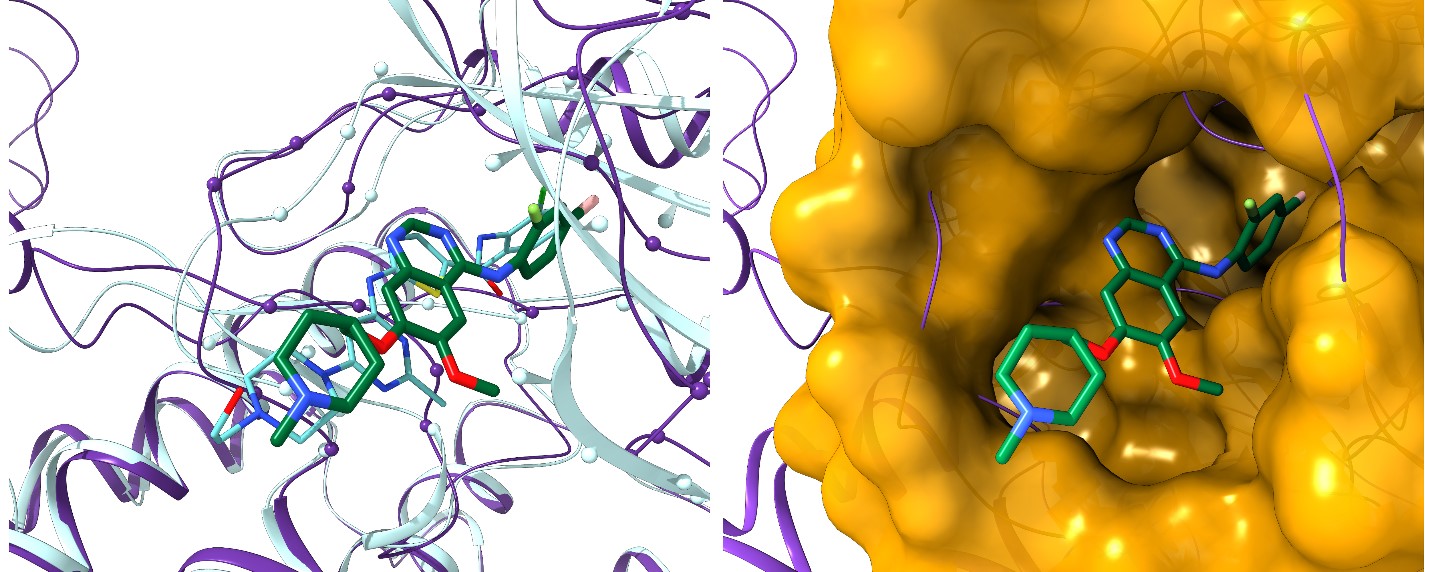
About 7,000 rare diseases currently affect more than 350 million people worldwide. Although these conditions collectively pose significant health care problems, commercial drug discovery for orphan diseases is financially complicated due to the limits of individual markets. Developing new treatments for diseases that fall into this category often requires financial incentives from governments, special research grants, and private philanthropy. To circumvent the economic hurdles, researchers have found drug repositioning to be a cheaper and faster alternative to traditional drug discovery. This method extends the purpose of a drug beyond its intended disease by identifying other conditions it can be effective against and significantly reducing research costs. Although drug repositioning is time effective, economically advantageous, and has great potential in the development of treatments for rare conditions, each repositioning application possibility comes with its own set of challenges. In order to mete out the complexities of these challenges, rational and effective computational methods to develop solutions for existing therapeutics are being explored. These pioneering computer simulations systematically evaluate possible links between currently FDA-approved drugs and potential therapeutic targets for rare diseases, allowing the most promising candidates for repositioning to be tested in the lab.
The Computational Systems Biology Group (CSBG) led by Dr. Michal Brylinski at Louisiana State University has developed a collection of tools for drug repositioning computer simulations. CSBG researchers used this promising methodology to match 715 known drugs against 922 potential drug targets implicated in rare diseases. Their results revealed as many as 18,145 candidates for repurposing, unveiling a number of opportunities to combat rare diseases with existing drugs. The team conducted their simulations on Louisiana Optical Network Infrastructure’s (LONI) Queen Bee 2 (QB2) supercomputer. Equipped with more than 10,000 CPU cores and more than 1,000 General Purpose Graphic Processing Units (GPGPUs), QB2 provides an ideal platform for researchers working on large-scale problems, such as computer-aided drug discovery and repositioning.
Since its inception more than a decade ago, LONI has provided cyberinfrastructure that is vital for researchers like CSBG to fulfill their missions and make noteworthy contributions to scientific research in the State of Louisiana. In 2018, QB2 supported a total of 168 research projects, representing a wide variety of scientific disciplines not limited to engineering, material sciences, chemistry, and biological sciences. During the year, 226 researchers from 12 institutions ran 132,848 jobs on QB2, consuming more than 50 million CPU-hours.